NGSEP
This is the web app or web related tool named NGSEP whose latest release can be downloaded as NGSEPplugin_3.3.0.201810051117.jar from this website redcoolmedia.net
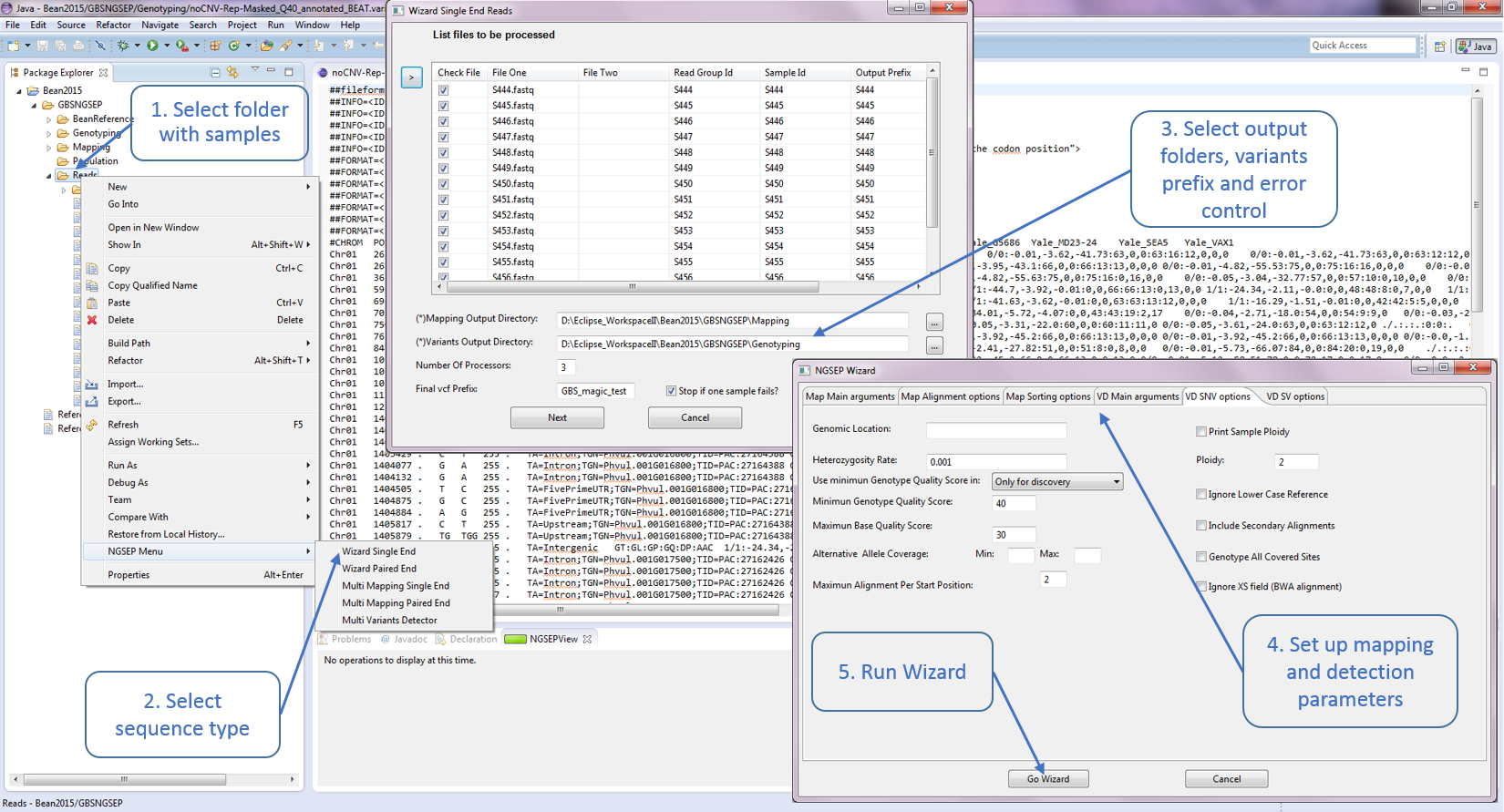
SCREENSHOTS:
NGSEP
APP DESCRIPTION:
Download this app named NGSEP.
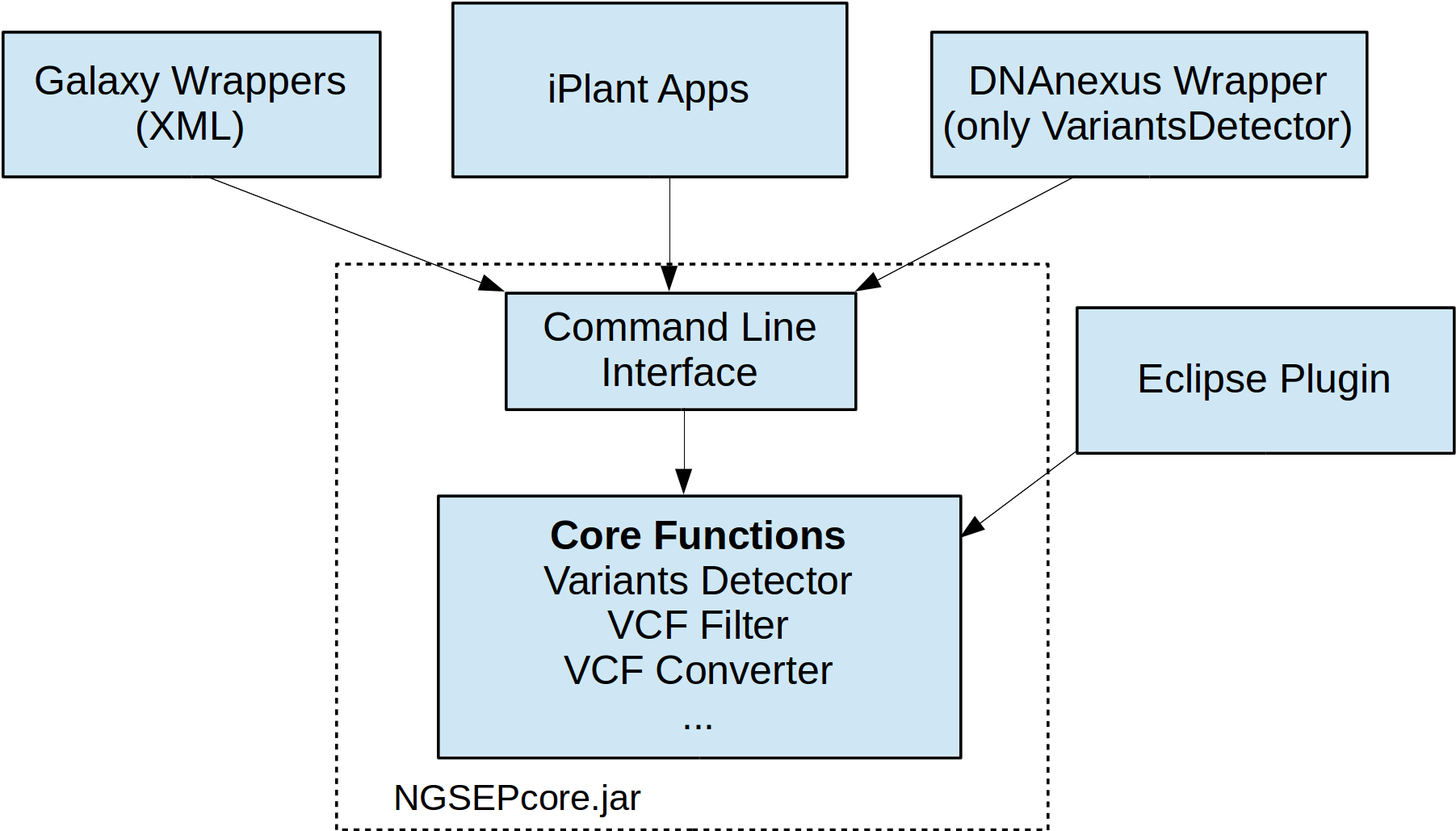
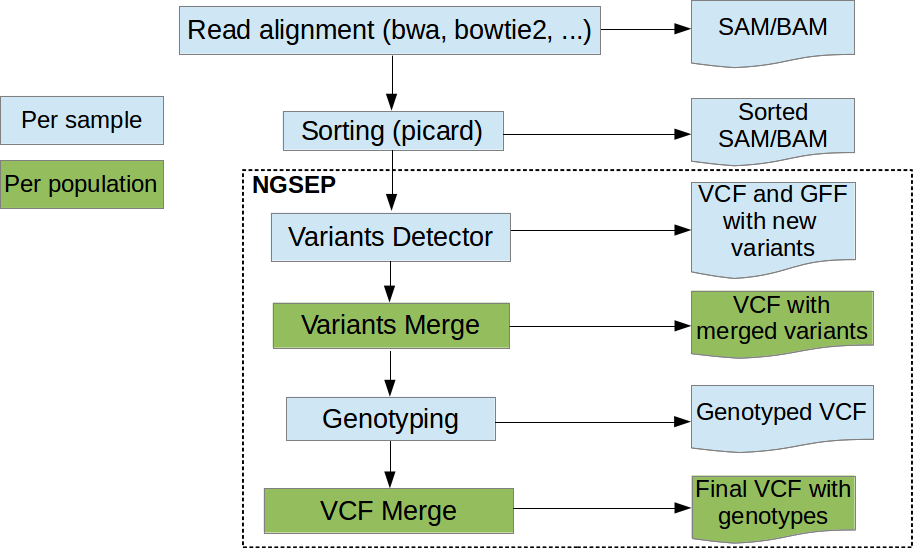
NGSEP is an integrated framework for analysis of DNA high throughput sequencing data. The main use of NGSEP is the construction and downstream analysis of large datasets of genomic variation. NGSEP performs accurate detection and genotyping of Single Nucleotide Variants (SNVs), small and large indels, short tandem repeats (STRs), inversions, and Copy Number Variants (CNVs). NGSEP also provides modules for functional annotation, filtering, format conversion, comparison, clustering, imputation, introgression analysis and different kinds of statistics. Other functionalities include calculations of k-mer distributions from fasta or fastq files, demultiplexing of barcoded sequencing reads, and comparative analysis of read depth distributions.Note: The quick download (green) button will provide you the jar file for command line usage. For other usage alternatives, please visit the wiki (https://sourceforge.net/p/ngsep/wiki/Home/)
Features
- SNPs, CNVs and Structural Variants detection
- Functional Annotation
- VCF manipulation: merge, filter, compare, format conversion
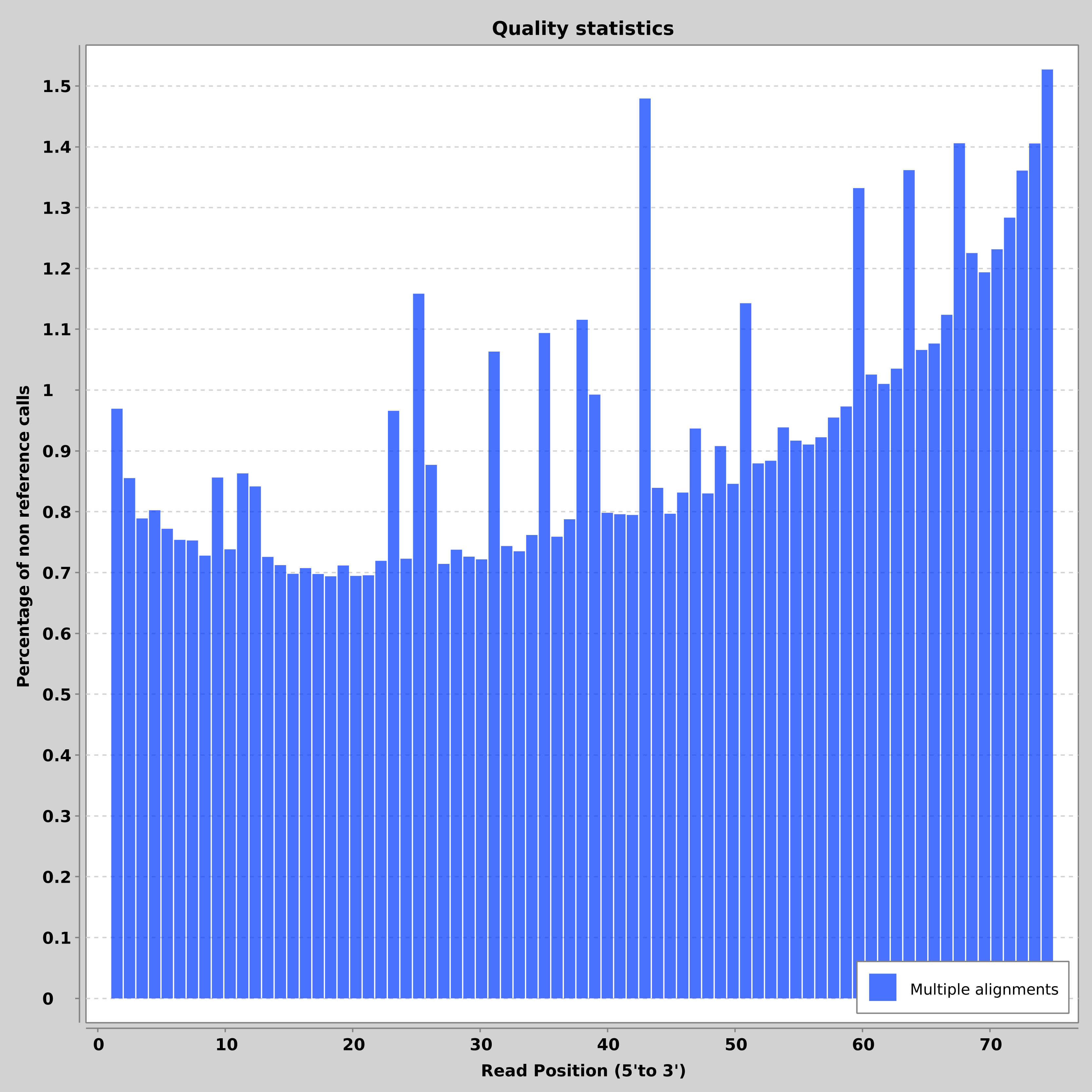
- SAM/BAM and VCF statistics calculation and plotting
- Reads Demultiplexing
- Genotype Imputation
- Graphical interface for Bowtie2 mapping
- Web Deployment through integration with Galaxy
- A graphical interface within the Cyverse collaborative platform
Audience
Healthcare Industry, Science/Research, Other Audience, Agriculture
User interface
Java SWT, Console/Terminal, Eclipse
Programming Language
Java
Free download Web app or web tool NGSEP from RedcoolMedia.net