Central Proteomics Facilities Pipeline
This is the web app or web related tool named Central Proteomics Facilities Pipeline whose latest release can be downloaded as README from this website redcoolmedia.net
SCREENSHOTS:
Central Proteomics Facilities Pipeline
APP DESCRIPTION:
Download this app named Central Proteomics Facilities Pipeline.
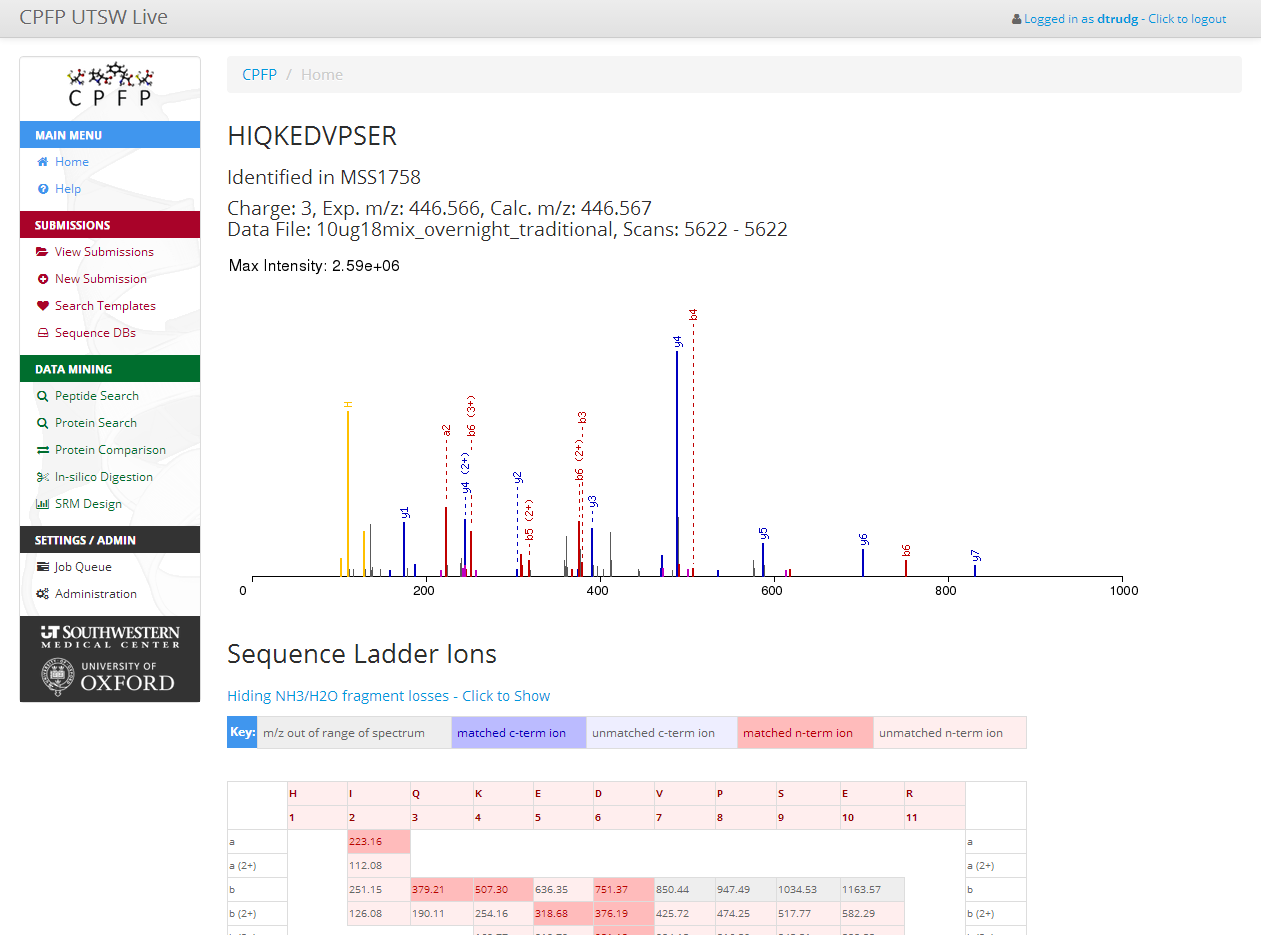
CPFP provides a pipeline for the analysis of MS/MS proteomic data, targeted at the needs of central proteomics facilities.== Project Status - 2014/08/14 ==
Due to the departure of the main developer from UTSW at the end of August 2014, major development of CPFP is unlikely in the immediate future. The software remains in use at UTSW and Oxford, where bug fixes and minor features continue to be implemented.
Potential users of CPFP are advised to consider the availability of local expertise and computing support before selecting CPFP as an analysis platform.
Please view the project website at cpfp.sf.net for full details and access to a demonstration server.
Please use the mailing list for any queries:
https://lists.sourceforge.net/lists/listinfo/cpfp-users
Features
- Web based proteomics pipeline targetted at needs of central proteomics facilities
- MS/MS Search with Mascot, X!Tandem, OMSSA in a single step
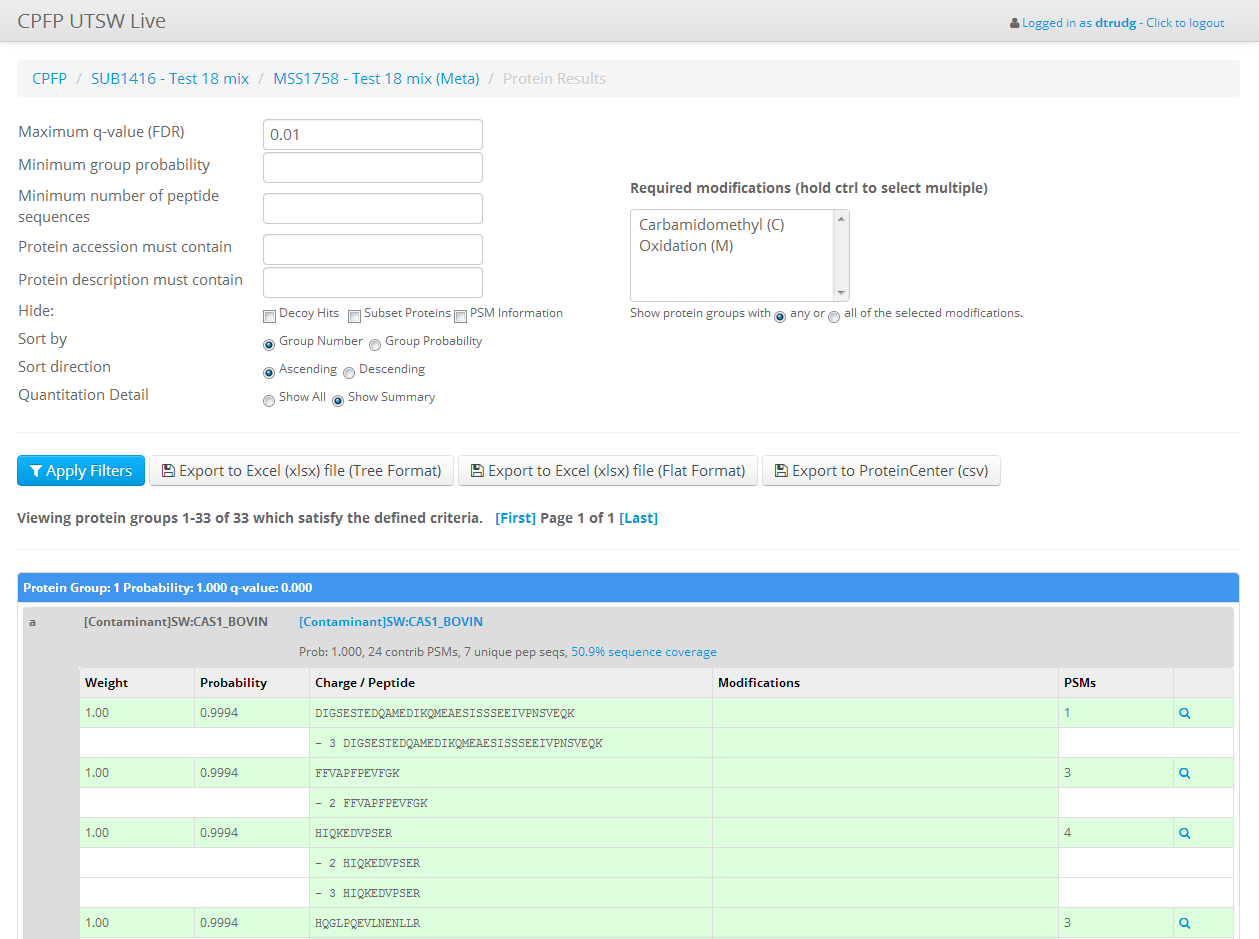
- Peptide and Protein validation using PeptideProphet & ProteinProphet
- Supports iTRAQ, SILAC, and SINQ Spectral Index label-free quantitation
Free download Web app or web tool Central Proteomics Facilities Pipeline from RedcoolMedia.net